Draw volcano plot.
volcano_plot(
object,
fc_column_name = "fc",
p_value_column_name = "p_value_adjust",
labs_x = "log2(Fold change)",
labs_y = "-log(p-adjust, 10)",
fc_up_cutoff = 2,
fc_down_cutoff = 0.5,
p_value_cutoff = 0.05,
line_color = "red",
up_color = "#EE0000FF",
down_color = "#3B4992FF",
no_color = "#808180FF",
point_size = 2,
point_alpha = 1,
point_size_scale,
line_type = 1,
add_text = FALSE,
text_for = c("marker", "UP", "DOWM"),
text_from = "variable_id"
)Arguments
- object
tidymass-class object.
- fc_column_name
fc_column_name
- p_value_column_name
p_value_column_name
- labs_x
labs_x
- labs_y
labs_y
- fc_up_cutoff
fc_up_cutoff
- fc_down_cutoff
fc_down_cutoff
- p_value_cutoff
p_value_cutoff
- line_color
line_color
- up_color
up_color
- down_color
down_color
- no_color
no_color
- point_size
point_size
- point_alpha
point_alpha
- point_size_scale
point_size_scale
- line_type
line_type
- add_text
add_text
- text_for
text_for
- text_from
text_from
Value
ggplot2 object
Examples
library(massdataset)
library(magrittr)
library(dplyr)
data("liver_aging_pos")
liver_aging_pos
#> --------------------
#> massdataset version: 0.01
#> --------------------
#> 1.expression_data:[ 21607 x 24 data.frame]
#> 2.sample_info:[ 24 x 4 data.frame]
#> 3.variable_info:[ 21607 x 3 data.frame]
#> 4.sample_info_note:[ 4 x 2 data.frame]
#> 5.variable_info_note:[ 3 x 2 data.frame]
#> 6.ms2_data:[ 0 variables x 0 MS2 spectra]
#> --------------------
#> Processing information (extract_process_info())
#> 1 processings in total
#> Creation ----------
#> Package Function.used Time
#> 1 massdataset create_mass_dataset() 2021-12-23 00:24:02
w_78 =
liver_aging_pos %>%
activate_mass_dataset(what = "sample_info") %>%
dplyr::filter(group == "78W") %>%
dplyr::pull(sample_id)
w_24 =
liver_aging_pos %>%
activate_mass_dataset(what = "sample_info") %>%
dplyr::filter(group == "24W") %>%
dplyr::pull(sample_id)
control_sample_id = w_24
case_sample_id = w_78
liver_aging_pos =
mutate_fc(
object = liver_aging_pos,
control_sample_id = control_sample_id,
case_sample_id = case_sample_id,
mean_median = "mean"
)
#> 10 control samples.
#> 10 case samples.
liver_aging_pos =
mutate_p_value(
object = liver_aging_pos,
control_sample_id = control_sample_id,
case_sample_id = case_sample_id,
method = "t.test",
p_adjust_methods = "BH"
)
#> 10 control samples.
#> 10 case samples.
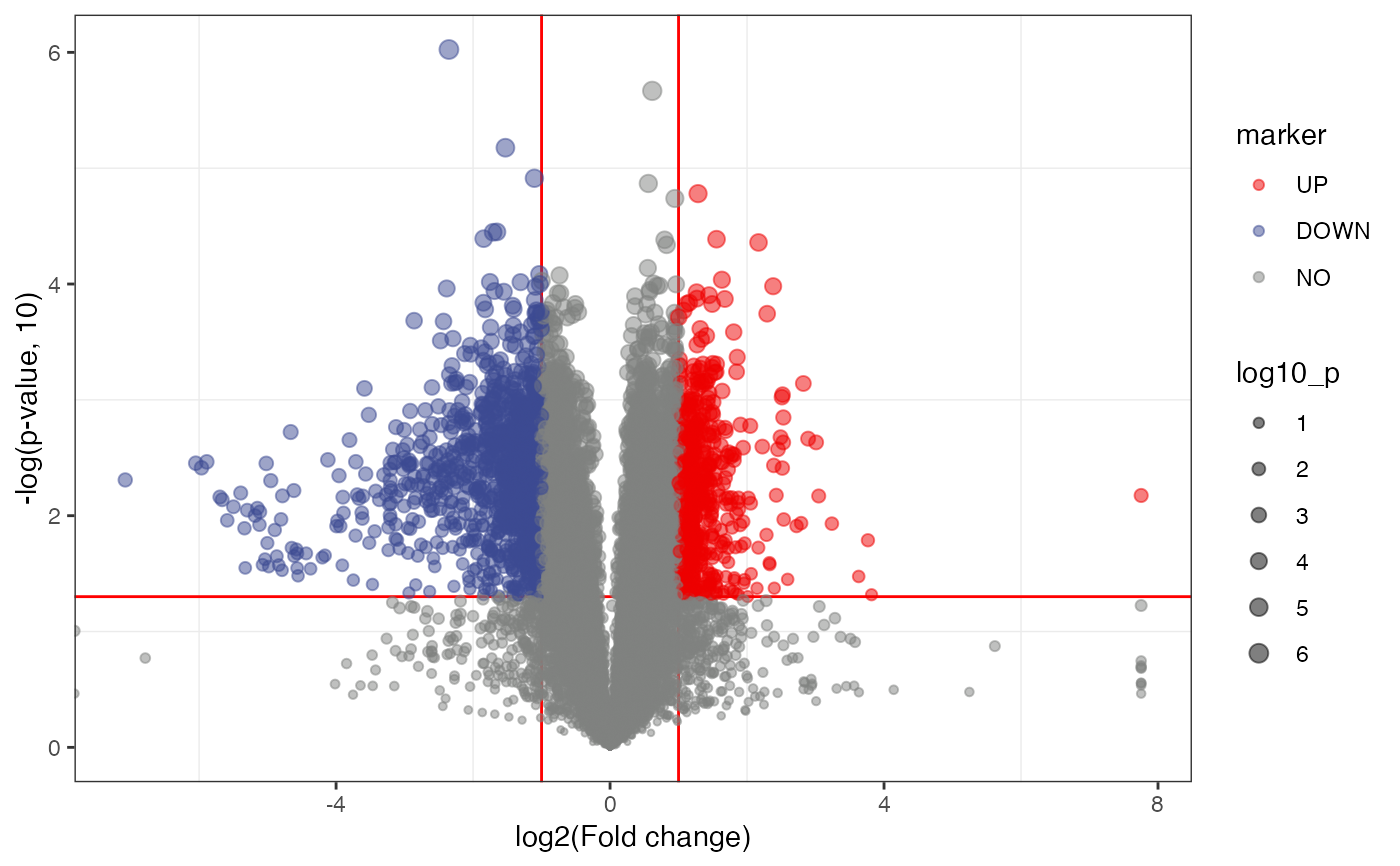
object = liver_aging_pos
volcano_plot(
object = object,
fc_column_name = "fc",
p_value_column_name = "p_value_adjust",
labs_x = "log2(Fold change)",
labs_y = "-log(p-adjust, 10)",
fc_up_cutoff = 2,
fc_down_cutoff = 0.5,
p_value_cutoff = 0.05,
add_text = TRUE
)
#> Warning: Removed 10 rows containing missing values (geom_point).
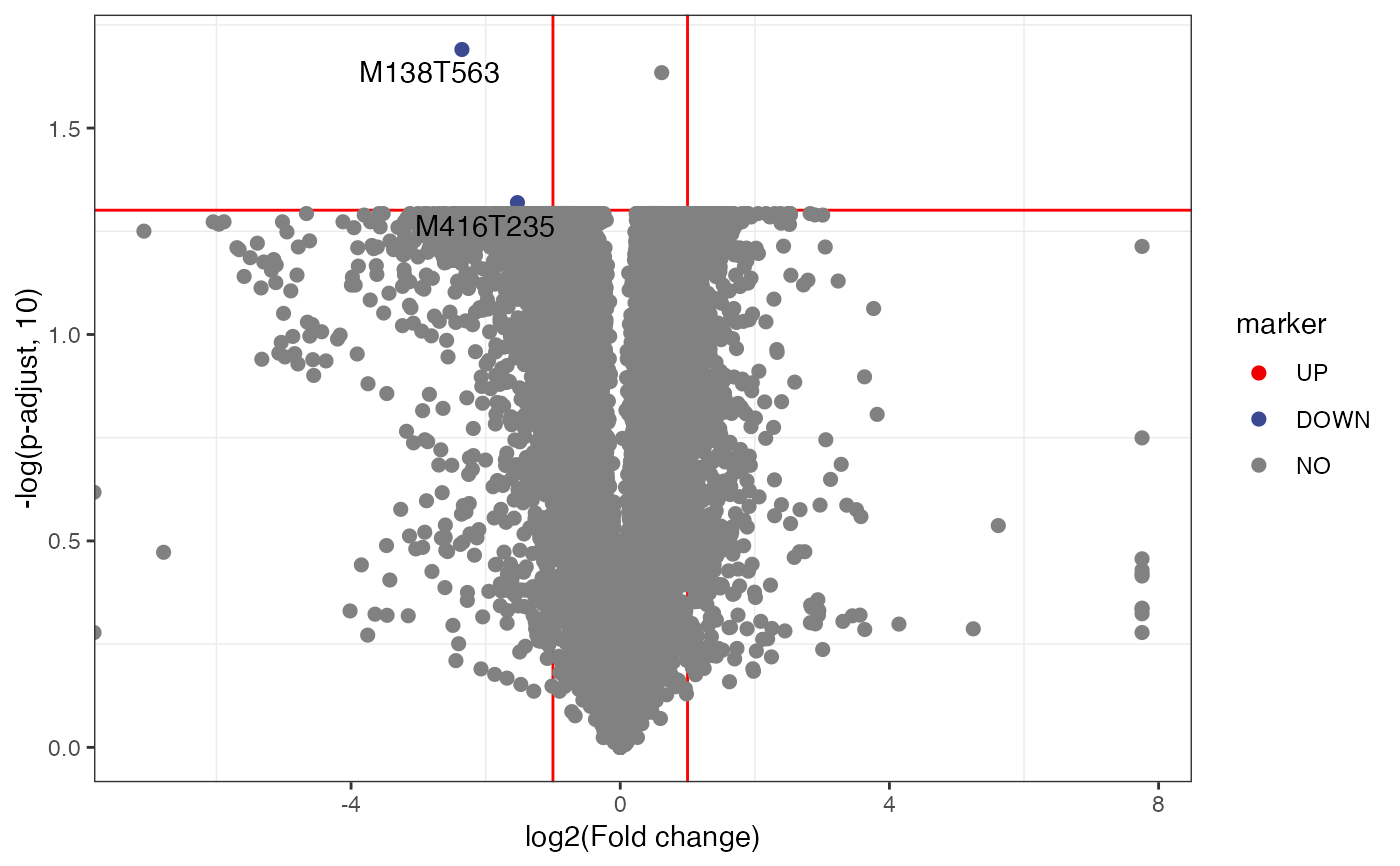 volcano_plot(
object = object,
fc_column_name = "fc",
p_value_column_name = "p_value",
labs_x = "log2(Fold change)",
labs_y = "-log(p-value, 10)",
fc_up_cutoff = 2,
fc_down_cutoff = 0.5,
p_value_cutoff = 0.05,
add_text = FALSE,
point_alpha = 0.5
)
#> Warning: Removed 10 rows containing missing values (geom_point).
volcano_plot(
object = object,
fc_column_name = "fc",
p_value_column_name = "p_value",
labs_x = "log2(Fold change)",
labs_y = "-log(p-value, 10)",
fc_up_cutoff = 2,
fc_down_cutoff = 0.5,
p_value_cutoff = 0.05,
add_text = FALSE,
point_alpha = 0.5
)
#> Warning: Removed 10 rows containing missing values (geom_point).
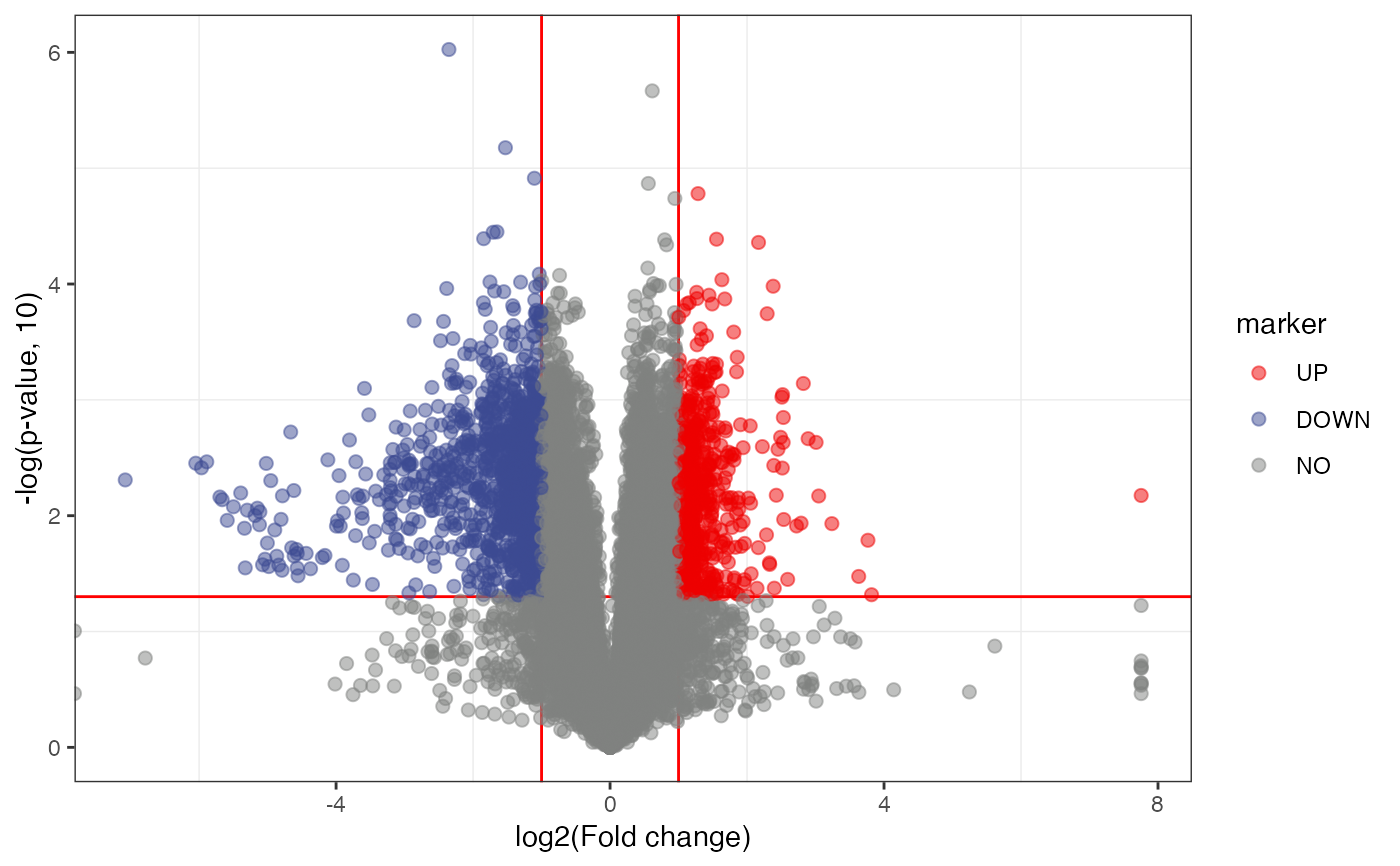 volcano_plot(
object = object,
fc_column_name = "fc",
p_value_column_name = "p_value",
labs_x = "log2(Fold change)",
labs_y = "-log(p-value, 10)",
fc_up_cutoff = 2,
fc_down_cutoff = 0.5,
p_value_cutoff = 0.05,
add_text = FALSE,
point_alpha = 0.5,
point_size_scale = "p_value"
) +
scale_size_continuous(range = c(0.5, 3))
#> Warning: Removed 10 rows containing missing values (geom_point).
volcano_plot(
object = object,
fc_column_name = "fc",
p_value_column_name = "p_value",
labs_x = "log2(Fold change)",
labs_y = "-log(p-value, 10)",
fc_up_cutoff = 2,
fc_down_cutoff = 0.5,
p_value_cutoff = 0.05,
add_text = FALSE,
point_alpha = 0.5,
point_size_scale = "p_value"
) +
scale_size_continuous(range = c(0.5, 3))
#> Warning: Removed 10 rows containing missing values (geom_point).