Distance and correlation
Xiaotao Shen (https://www.shenxt.info/)
Created on 2022-02-18 and updated on 2022-09-19
Source:vignettes/dist_cor.Rmd
dist_cor.RmdData preparation
library(massdataset)
library(tidyverse)
library(massstat)
data("liver_aging_pos")
liver_aging_pos
#> --------------------
#> massdataset version: 0.01
#> --------------------
#> 1.expression_data:[ 21607 x 24 data.frame]
#> 2.sample_info:[ 24 x 4 data.frame]
#> 3.variable_info:[ 21607 x 3 data.frame]
#> 4.sample_info_note:[ 4 x 2 data.frame]
#> 5.variable_info_note:[ 3 x 2 data.frame]
#> 6.ms2_data:[ 0 variables x 0 MS2 spectra]
#> --------------------
#> Processing information (extract_process_info())
#> Creation ----------
#> Package Function.used Time
#> 1 massdataset create_mass_dataset() 2021-12-23 00:24:02Distance
library(massdataset)
library(tidyverse)
data("expression_data")
data("sample_info")
data("sample_info_note")
data("variable_info")
data("variable_info_note")
object =
create_mass_dataset(
expression_data = expression_data,
sample_info = sample_info,
variable_info = variable_info,
sample_info_note = sample_info_note,
variable_info_note = variable_info_note
)
object
#> --------------------
#> massdataset version: 0.99.6
#> --------------------
#> 1.expression_data:[ 1000 x 8 data.frame]
#> 2.sample_info:[ 8 x 4 data.frame]
#> 3.variable_info:[ 1000 x 3 data.frame]
#> 4.sample_info_note:[ 4 x 2 data.frame]
#> 5.variable_info_note:[ 3 x 2 data.frame]
#> 6.ms2_data:[ 0 variables x 0 MS2 spectra]
#> --------------------
#> Processing information (extract_process_info())
#> create_mass_dataset ----------
#> Package Function.used Time
#> 1 massdataset create_mass_dataset() 2022-02-18 12:46:21
x =
object %>%
log(2) %>%
scale()
variable_distance <-
dist_mass_dataset(x = x, margin = "variable")
head(as.matrix(variable_distance)[, 1:5])
#> M136T55_2_POS M79T35_POS M307T548_POS M183T224_POS M349T47_POS
#> M136T55_2_POS 0.000000 2.595449 4.743621 4.708308 1.940902
#> M79T35_POS 2.595449 0.000000 4.219588 3.106812 1.620770
#> M307T548_POS 4.743621 4.219588 0.000000 2.568996 3.986112
#> M183T224_POS 4.708308 3.106812 2.568996 0.000000 3.082528
#> M349T47_POS 1.940902 1.620770 3.986112 3.082528 0.000000
#> M182T828_POS 3.010099 2.743352 3.139591 2.175595 2.895630
sample_distance <-
dist_mass_dataset(x = x, margin = "sample")
head(as.matrix(sample_distance)[, 1:5])
#> Blank_3 Blank_4 QC_1 QC_2 PS4P1
#> Blank_3 0.00000 46.24363 46.22024 54.26607 47.31000
#> Blank_4 46.24363 0.00000 53.89434 60.88273 55.83349
#> QC_1 46.22024 53.89434 0.00000 40.44652 39.65971
#> QC_2 54.26607 60.88273 40.44652 0.00000 46.26019
#> PS4P1 47.31000 55.83349 39.65971 46.26019 0.00000
#> PS4P2 47.06702 55.78646 38.33452 49.76545 39.14705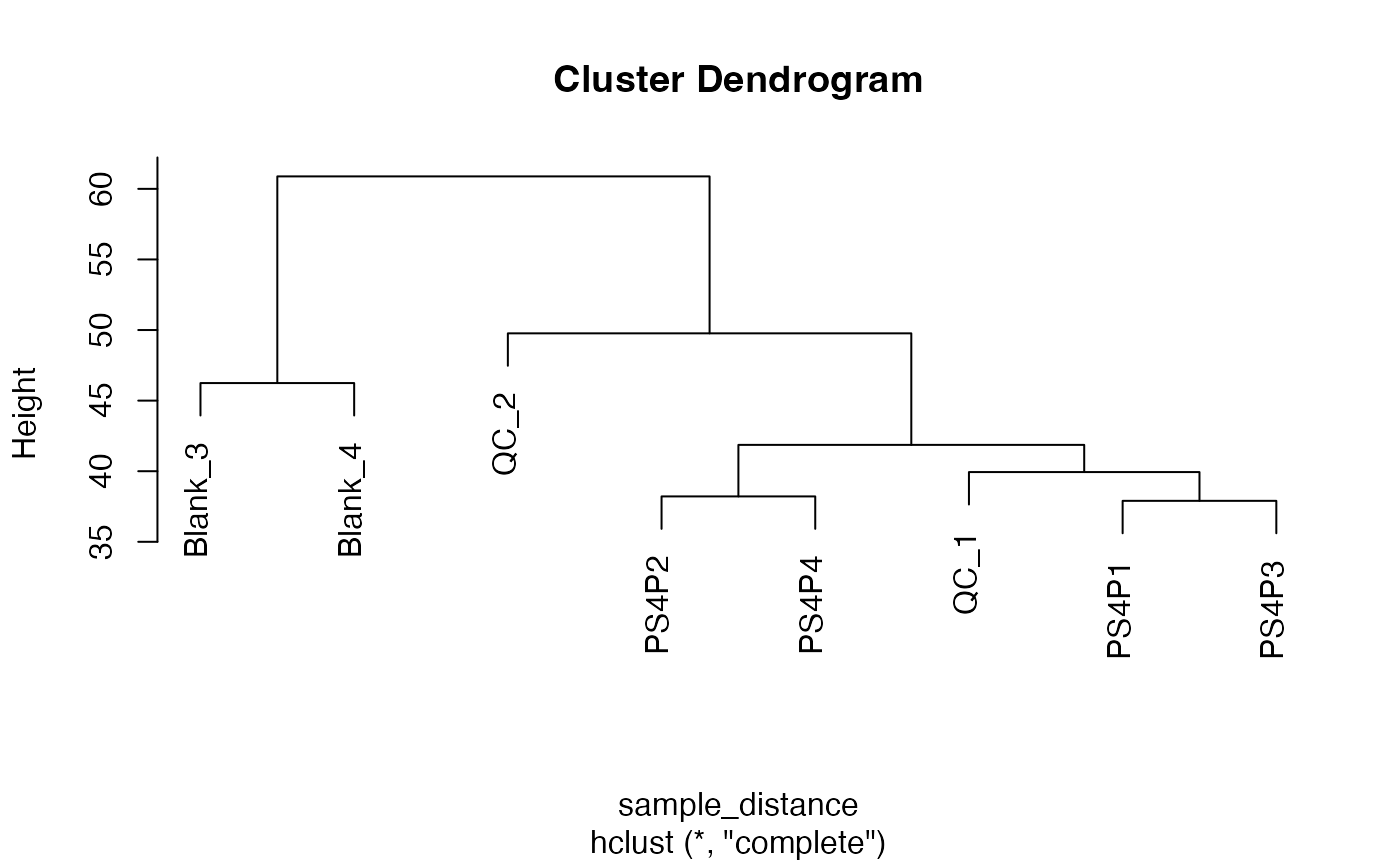
Correlation
library(massdataset)
library(tidyverse)
data("liver_aging_pos")
x =
liver_aging_pos %>%
`+`(1) %>%
log(2) %>%
scale()
variable_cor <-
cor_mass_dataset(x = x[1:100,], margin = "variable")
head(as.matrix(variable_cor)[, 1:5])
#> M60T1209 M60T1182 M60T1163 M60T1310 M60T1240
#> M60T1209 1.00000000 -0.279569382 -0.131480430 0.04905720 0.04196437
#> M60T1182 -0.27956938 1.000000000 0.001388209 -0.28540425 -0.16890419
#> M60T1163 -0.13148043 0.001388209 1.000000000 -0.08866161 0.07438262
#> M60T1310 0.04905720 -0.285404246 -0.088661609 1.00000000 -0.21471600
#> M60T1240 0.04196437 -0.168904192 0.074382619 -0.21471600 1.00000000
#> M60T1254 0.03039983 -0.300416254 0.246969867 -0.02536577 0.53425798
sample_cor <-
cor_mass_dataset(x = x, margin = "sample")
head(as.matrix(sample_cor)[, 1:5])
#> QC01 QC02 QC03 QC04 liver24_10
#> QC01 1.000000000 -0.2111021 -0.20233982 -0.2335868 -0.003668197
#> QC02 -0.211102150 1.0000000 0.74699358 0.7539063 -0.248487135
#> QC03 -0.202339821 0.7469936 1.00000000 0.7680612 -0.263692199
#> QC04 -0.233586820 0.7539063 0.76806121 1.0000000 -0.254396152
#> liver24_10 -0.003668197 -0.2484871 -0.26369220 -0.2543962 1.000000000
#> liver24_1 0.003550632 -0.1169218 -0.07114657 -0.1150889 -0.222070187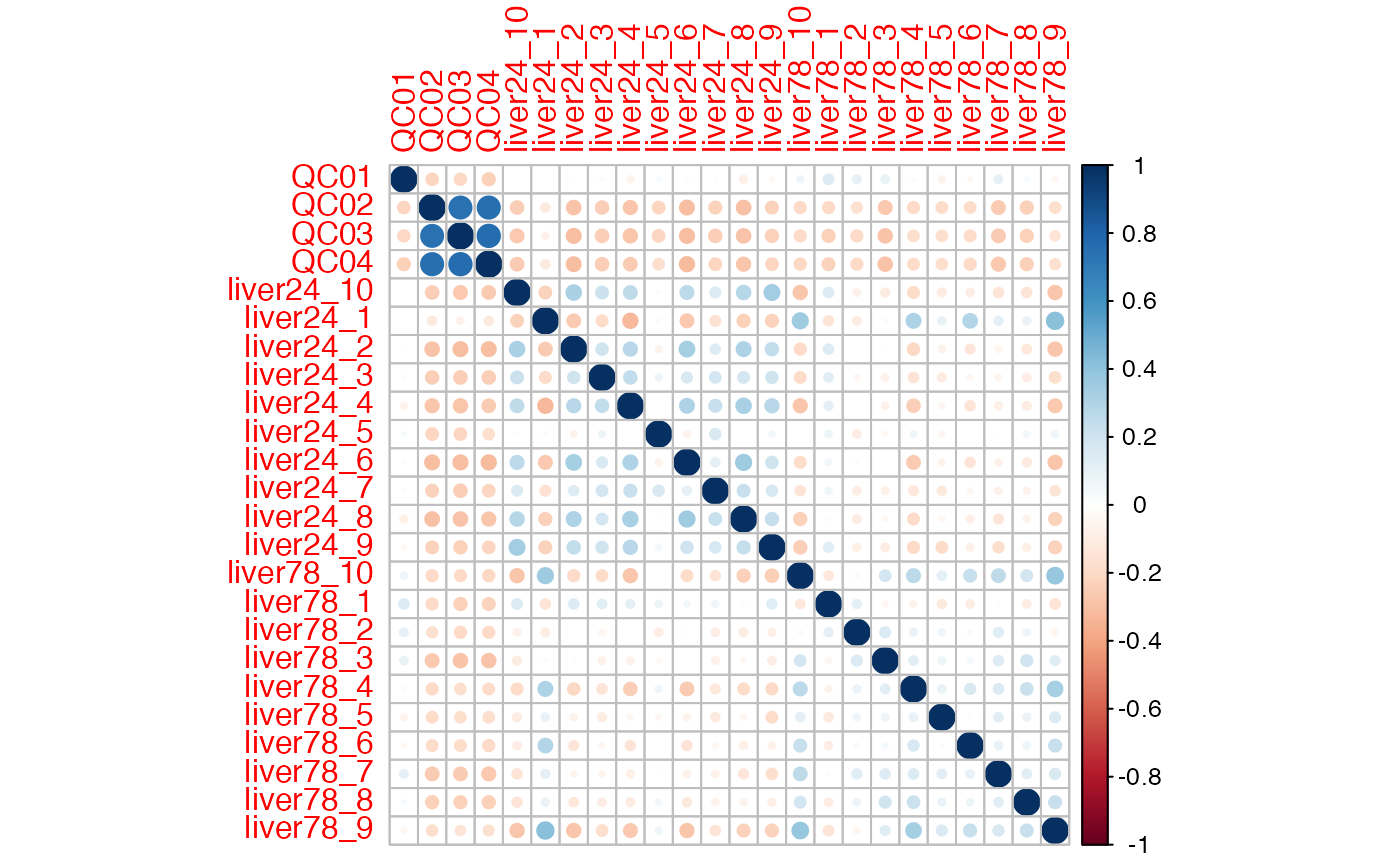
Session information
sessionInfo()
#> R version 4.1.2 (2021-11-01)
#> Platform: x86_64-apple-darwin17.0 (64-bit)
#> Running under: macOS Big Sur 10.16
#>
#> Matrix products: default
#> BLAS: /Library/Frameworks/R.framework/Versions/4.1/Resources/lib/libRblas.0.dylib
#> LAPACK: /Library/Frameworks/R.framework/Versions/4.1/Resources/lib/libRlapack.dylib
#>
#> locale:
#> [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] corrplot_0.92 ggfortify_0.4.14 massstat_0.99.4 forcats_0.5.1
#> [5] stringr_1.4.0 dplyr_1.0.7 purrr_0.3.4 readr_2.1.1
#> [9] tidyr_1.1.4 tibble_3.1.6 ggplot2_3.3.5 tidyverse_1.3.1
#> [13] magrittr_2.0.2 masstools_0.99.3 massdataset_0.99.6
#>
#> loaded via a namespace (and not attached):
#> [1] readxl_1.3.1 snow_0.4-4 backports_1.4.1
#> [4] circlize_0.4.14 systemfonts_1.0.3 igraph_1.2.11
#> [7] plyr_1.8.6 lazyeval_0.2.2 BiocParallel_1.28.3
#> [10] crosstalk_1.2.0 leaflet_2.0.4.1 digest_0.6.29
#> [13] foreach_1.5.1 yulab.utils_0.0.4 htmltools_0.5.2
#> [16] fansi_1.0.2 memoise_2.0.1 cluster_2.1.2
#> [19] doParallel_1.0.16 tzdb_0.2.0 openxlsx_4.2.5
#> [22] limma_3.50.0 ComplexHeatmap_2.10.0 modelr_0.1.8
#> [25] matrixStats_0.61.0 rARPACK_0.11-0 pkgdown_2.0.2
#> [28] colorspace_2.0-2 rvest_1.0.2 ggrepel_0.9.1
#> [31] textshaping_0.3.6 haven_2.4.3 xfun_0.29
#> [34] crayon_1.4.2 jsonlite_1.7.3 impute_1.68.0
#> [37] iterators_1.0.13 glue_1.6.1 gtable_0.3.0
#> [40] zlibbioc_1.40.0 GetoptLong_1.0.5 shape_1.4.6
#> [43] BiocGenerics_0.40.0 scales_1.1.1 vsn_3.62.0
#> [46] DBI_1.1.2 Rcpp_1.0.8 mzR_2.28.0
#> [49] viridisLite_0.4.0 clue_0.3-60 gridGraphics_0.5-1
#> [52] preprocessCore_1.56.0 clisymbols_1.2.0 stats4_4.1.2
#> [55] MsCoreUtils_1.6.0 htmlwidgets_1.5.4 httr_1.4.2
#> [58] RColorBrewer_1.1-2 ellipsis_0.3.2 pkgconfig_2.0.3
#> [61] XML_3.99-0.8 sass_0.4.0 dbplyr_2.1.1
#> [64] utf8_1.2.2 reshape2_1.4.4 ggplotify_0.1.0
#> [67] tidyselect_1.1.1 rlang_1.0.0 munsell_0.5.0
#> [70] cellranger_1.1.0 tools_4.1.2 cachem_1.0.6
#> [73] cli_3.1.1 generics_0.1.1 broom_0.7.12
#> [76] evaluate_0.14 fastmap_1.1.0 mzID_1.32.0
#> [79] yaml_2.2.2 ragg_1.2.1 knitr_1.37
#> [82] fs_1.5.2 zip_2.2.0 ncdf4_1.19
#> [85] pbapply_1.5-0 xml2_1.3.3 compiler_4.1.2
#> [88] rstudioapi_0.13 plotly_4.10.0 png_0.1-7
#> [91] affyio_1.64.0 reprex_2.0.1 bslib_0.3.1
#> [94] stringi_1.7.6 highr_0.9 RSpectra_0.16-0
#> [97] desc_1.4.0 MSnbase_2.20.4 lattice_0.20-45
#> [100] Matrix_1.4-0 ProtGenerics_1.26.0 ggsci_2.9
#> [103] vctrs_0.3.8 pillar_1.6.5 lifecycle_1.0.1
#> [106] BiocManager_1.30.16 jquerylib_0.1.4 MALDIquant_1.21
#> [109] GlobalOptions_0.1.2 data.table_1.14.2 corpcor_1.6.10
#> [112] patchwork_1.1.1 R6_2.5.1 pcaMethods_1.86.0
#> [115] affy_1.72.0 gridExtra_2.3 IRanges_2.28.0
#> [118] codetools_0.2-18 fastDummies_1.6.3 MASS_7.3-55
#> [121] assertthat_0.2.1 rprojroot_2.0.2 rjson_0.2.21
#> [124] withr_2.4.3 S4Vectors_0.32.3 parallel_4.1.2
#> [127] hms_1.1.1 mixOmics_6.18.1 grid_4.1.2
#> [130] rmarkdown_2.11 Biobase_2.54.0 lubridate_1.8.0
#> [133] ellipse_0.4.2