pca_score_plot
pca_score_plot(
object,
pca_object,
x = 1,
y = 2,
color_by,
point_alpha = 1,
point_size = 5,
point_shape = 21,
frame = TRUE,
frame.type = "norm",
...
)Arguments
- object
mass_dataset.
- pca_object
pca_object from run_pca()
- x
PC x for X axis, default is 1.
- y
PC y for Y axis, default is 2.
- color_by
which column (sample_info) is used to color samples
- point_alpha
point_alpha default is 1.
- point_size
point_size, default is 5.
- point_shape
point_shape default is 21.
- frame
?ggplot2::autoplot
- frame.type
?ggplot2::autoplot
- ...
other paramters for ggplot2::autoplot
Value
ggplot2 plot.
Examples
library(massdataset)
library(magrittr)
library(dplyr)
data("liver_aging_pos")
liver_aging_pos
#> --------------------
#> massdataset version: 0.01
#> --------------------
#> 1.expression_data:[ 21607 x 24 data.frame]
#> 2.sample_info:[ 24 x 4 data.frame]
#> 3.variable_info:[ 21607 x 3 data.frame]
#> 4.sample_info_note:[ 4 x 2 data.frame]
#> 5.variable_info_note:[ 3 x 2 data.frame]
#> 6.ms2_data:[ 0 variables x 0 MS2 spectra]
#> --------------------
#> Processing information (extract_process_info())
#> 1 processings in total
#> Creation ----------
#> Package Function.used Time
#> 1 massdataset create_mass_dataset() 2021-12-23 00:24:02
pca_object =
liver_aging_pos %>%
scale() %>%
run_pca()
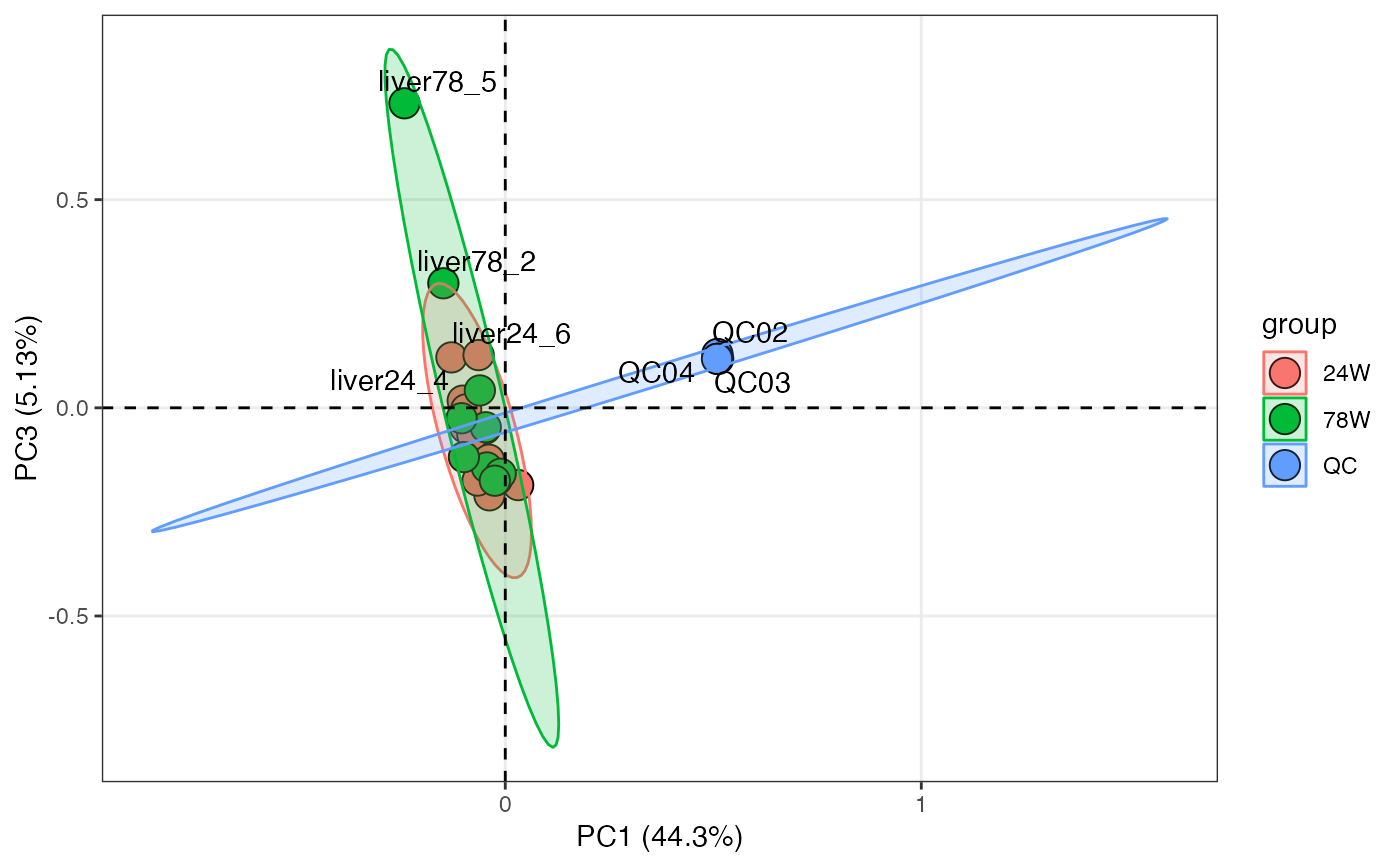
pca_score_plot(liver_aging_pos,
pca_object,
color_by = "group",
loadings = TRUE)
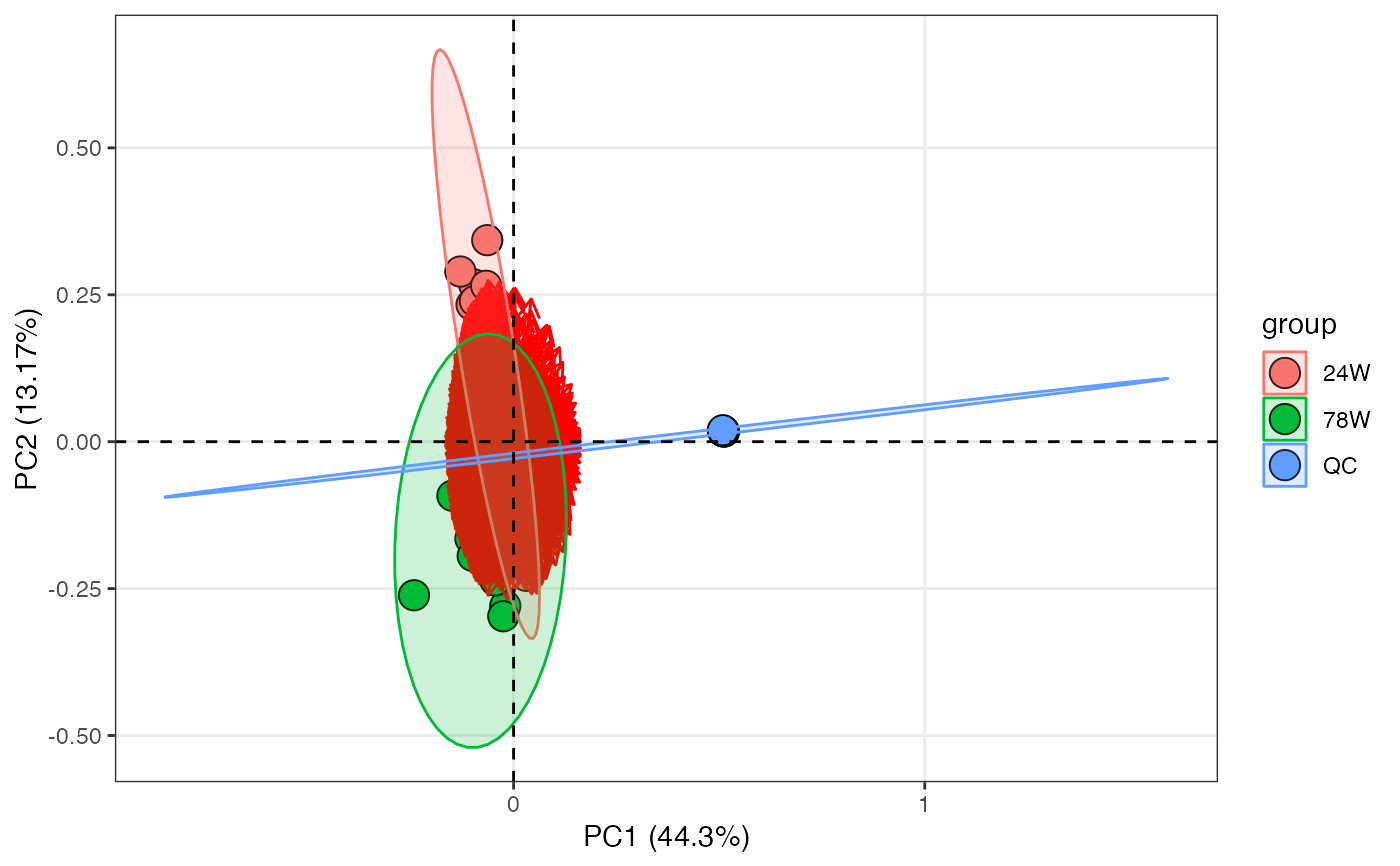 pca_score_plot(liver_aging_pos,
pca_object,
color_by = "group",
frame = TRUE) +
ggrepel::geom_text_repel(aes(label = sample_id))
pca_score_plot(liver_aging_pos,
pca_object,
color_by = "group",
frame = TRUE) +
ggrepel::geom_text_repel(aes(label = sample_id))
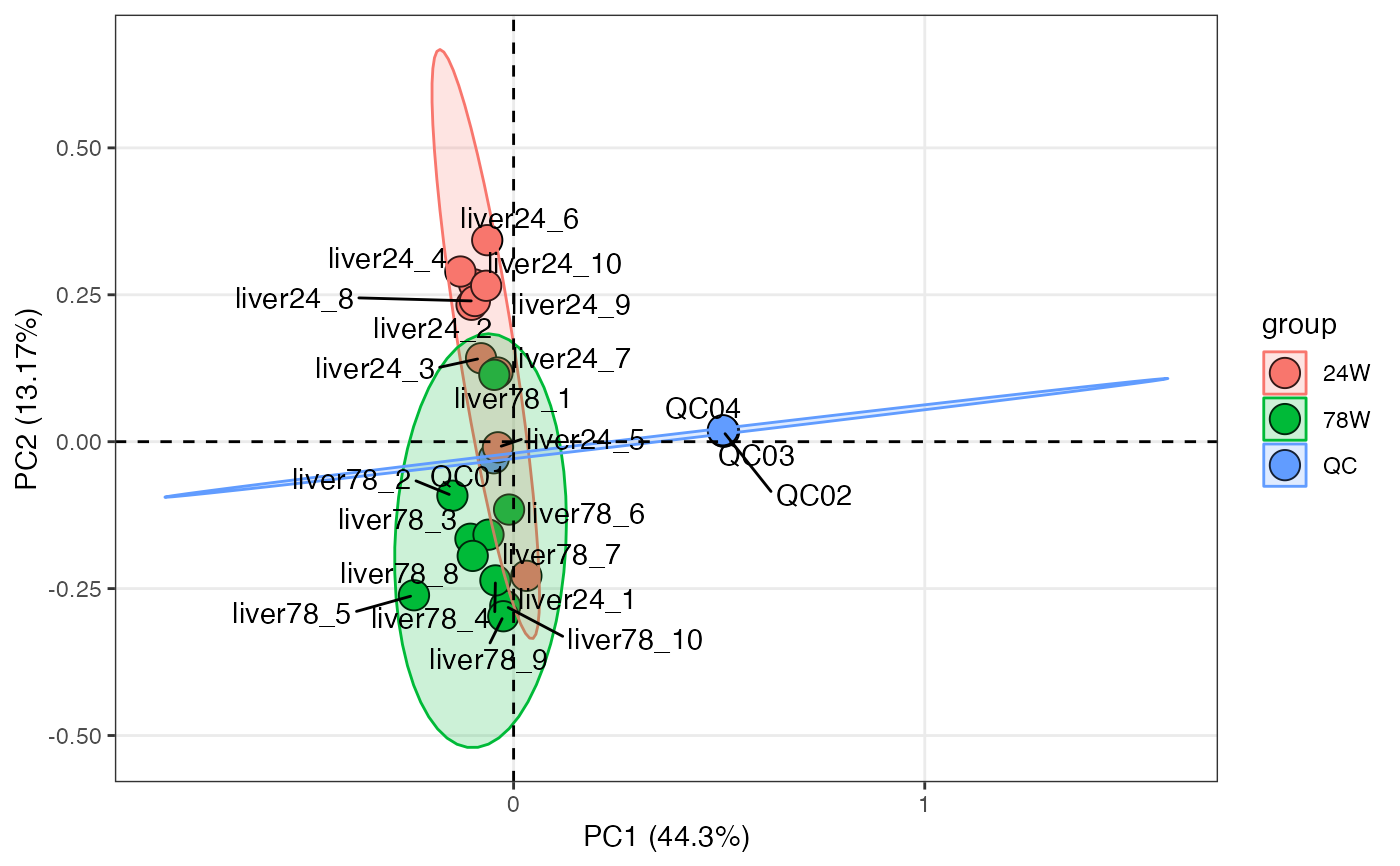 pca_score_plot(liver_aging_pos,
pca_object,
x = 1,
y = 3,
color_by = "group",
frame = TRUE) +
ggrepel::geom_text_repel(aes(label = sample_id))
#> Warning: ggrepel: 9 unlabeled data points (too many overlaps). Consider increasing max.overlaps
pca_score_plot(liver_aging_pos,
pca_object,
x = 1,
y = 3,
color_by = "group",
frame = TRUE) +
ggrepel::geom_text_repel(aes(label = sample_id))
#> Warning: ggrepel: 9 unlabeled data points (too many overlaps). Consider increasing max.overlaps